- Lentivirus
- Adeno-Associated
- Adenovirus
- Pseudovirus
- Vector
- Synthesis
- Autophagy Research
- CRISPR/Cas9
- Noncoding RNA
- Luciferase Assay
- Reagents
WHAT ARE YOU LOOKING FOR?
Luciferase is a general term for enzymes that can produce bioluminescence in nature, with the most representative ones are F-Luciferase from fireflies and R-Luciferase from renilla. Under the catalysis of luciferase, luciferin can efficiently convert to oxidized luciferin and emit strong fluorescence at the same time. Therefore, researchers used luciferase to develop a highly sensitive and convenient gene reporter system. Currently, this system is widely applied in various fields, including transcription factor binding site and promoter activity analysis, RNA degradation, signal transduction, drug screening, and in vivo animal imaging.
Classification | Service Content |
Transcription factor and promoter research | Promoter activity verification |
Interaction sites between transcription factors and promoters research | |
The interaction between miRNA and target genes research | Verification of miRNA target genes |
Verification of lncRNA-mediated miRNA sequestration | |
Verification of circRNA-mediated miRNA sequestration | |
Research on ceRNA mechanism |
Expert Academic Support: As a leading vector manufacturing company, Hanbio provides strong academic support team to assist in designing scientifically reasonable experimental schemes.
Rapid Vector Construction: The simple HB-infusion seamless cloning strategy greatly reduces the construction period of vectors.
High-Efficiency Transfection: The efficient LipoFiter transfection reagent and virus transduction system ensure stable and reproducible results.
Comprehensive Data Analysis: Detailed raw data and experimental result analysis guarantee clear and reliable conclusions with Hanbio's genetic engineering research.
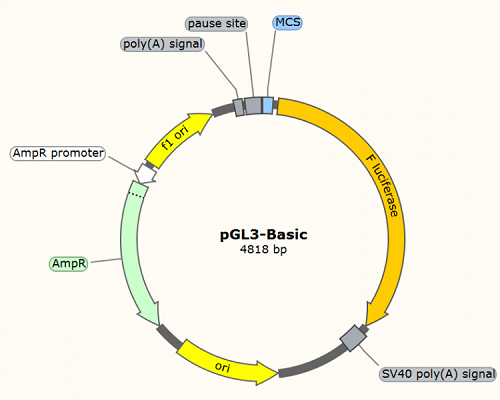
pGL3-Basic: used for transcription factor binding site and promoter activity analysis. The promoter region of interest and co-transfection with the transcription factor allows analysis of F-Luciferase activity, reflecting the activity level of the promoter. Typically, this plasmid requires combination with a constitutively expressing R-Luciferase vector as an internal control to normalize transfection efficiency between different samples (refer to Case1).
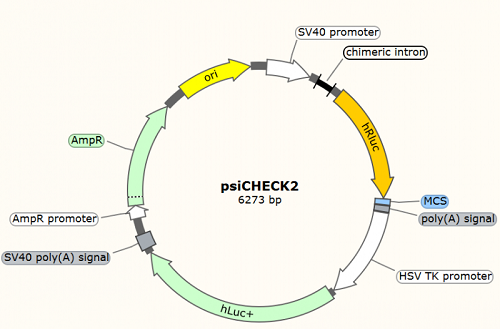
psiCHECK2: analysis of miRNA-target interactions, including miRNA target gene validation (refer to figure A and B of Case2), verification of miRNA sequestration by lncRNA (refer to Case3) and circRNA (refer to Case4). The potential miRNA binding sites are cloned into the 3'UTR region of R-Luciferase (hRLuc), and co-transfection with the miRNA of interest to determine the activity of R-Luciferase. F-Luciferase (hLuc+) is used as an internal control to normalize transfection efficiency between different samples.
Technical Scheme
Determining the experimental design based on research objectives
Constructing a Luciferase reporter gene system
Transfecting target cells
Measuring fluorescence activity
Reporting data
Classic Case1
Analysis of transcription factor binding sites and LncRNA promoter activity
Paper: Wang, P., et al. (2014). "The STAT3-binding long noncoding RNA lnc-DC controls human dendritic cell differentiation." Science 344(6181): 310-313.
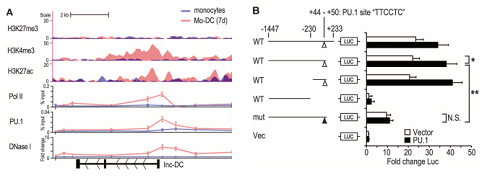
This article is a research paper published by the Caoxue Tao Lab. on Science in 2014. A lncRNA related to the development of dendritic cells (DC) was identified by microarray, named lnc-DC. High-throughput sequencing also found this differentially expressed lncRNA and then verified lnc-DC by Northern Blot. As lnc-DC is highly specifically and stably expressed during a certain period of DC development, the authors hypothesized that lnc-DC might be related to epigenetic regulation events. To verify this hypothesis, the author found that proteins such as Pol II, H3K4me3 and H3K27ac were enriched near the transcription start site (TSS) of lnc-DC by CHIP-seq and CHIP-qPCR (Figure A), suggesting that epigenetic changes during DC development correspond to transcriptional enhancement of lnc-DCs. Further sequence analysis revealed a classic PU.1 binding site near the TSS of lnc-DC (+44-+50). The authors hypothesized that PU.1 was involved in the specific expression of lnc-DC, so the following experiment was designed for further verification (Figure B).
The authors cloned the promoter region (-1447-+223) of lnc-DC and its different mutant into the upstream of the F-Luciferase expression box, and then co-transfected with the transcription factor PU.1 expression plasmid or the empty vector into the model cell line HEK-293T. The fluorescence intensity indirectly reflects the level of transcriptional activity. Finally, it was proved that the transcription factor PU.1 mediated the specific expression of lnc-DC during DC development through the canonical binding site (+44 - +50).
Classic Case2
LncRNA regulates the expression of target gene mRNA by adsorbing miRNA (ceRNA)
Paper: Cesana, M., et al. (2011). "A long noncoding RNA controls muscle differentiation by functioning as a competing endogenous RNA." Cell 147(2): 358-369.
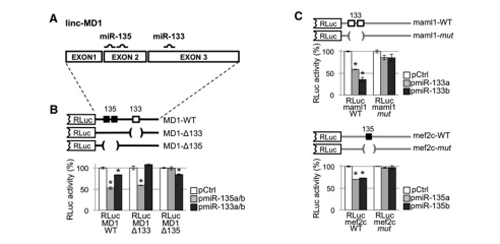
The authors identified a muscle development-related lncRNA, lnc-MD1, through expression analysis. Previous functional studies revealed that lnc-MD1 regulates the timing of muscle differentiation. Bioinformatics analysis identified 36 miRNA binding sites on this muscle-specific lncRNA. Considering only muscle development-related miRNAs and genes, miR-133 and miR-135 were the remaining candidates among the 36 miRNAs. The authors performed luciferase assays for validation. Firstly, they cloned lnc-MD1 wild-type and mutants lacking miR-133 and miR-135 binding sites into the 3'UTR of R-Luc, and then co-transfected with plasmids expressing miR-133 and miR-135 precursor into cells for luciferase activity detection (Figure A and B). It was proved that the binding sites of miR-133 and miR-135 existed in lnc-MD1.
Further bioinformatics predictions revealed that miR-133 and miR-135 target two genes, MAML1 and MEF2C, respectively. Similarly, the authors verified it by luciferase experiment. The 3'UTR of MAML1 and MEF2C and their mutants with lacking miRNA binding site were cloned downstream of the RLuc expression box, respectively. Then, they co-transfected them with miRNA overexpression vectors into cells for enzyme activity measurement. The results showed that miR-133 and miR-135 indeed targeted MAML1 and MEF2C genes respectively (Figure C). The final model is that lnc-MD1 absorbs miR-133 and 135 through the ceRNA mechanism, which positively regulates the expression of MAML1 and MEF2C, and participates in the regulation of muscle development.
Classic Case3
LncRNA regulates its physiological functions by sequestering miRNA (ceRNA)
Paper: Kallen, A. N., et al. (2013). "The imprinted H19 lncRNA antagonizes let-7 microRNAs." Molecular Cell 52(1): 101-112.
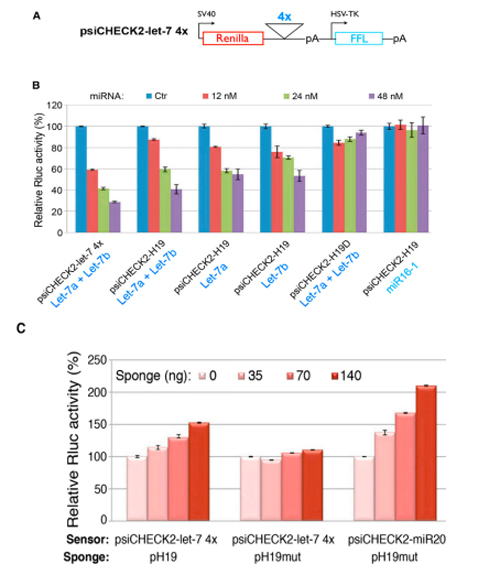
H19 belongs to a highly conserved imprinted gene family and plays a crucial role in embryonic development and growth. To further investigate the function of the lncRNA H19, the authors conducted bioinformatics analysis and identified four binding sites for let-7 within H19. Therefore, they hypothesized that H19 may affect the function of let-7.
To validate this prediction, the researchers cloned the four potential let-7 binding sites in tandem into the 3'UTR region of RLuc. Subsequently, they co-transfected with either a let-7 inhibitor vector (Figure A and C) or an overexpression vector (Figure B) into cell lines to measure luciferase activity, which confirmed that the binding site of Let-7 did exist and function. It was demonstrated that H19 participates in the regulation of muscle development by affecting the expression of let-7 family target genes through the mechanism of RNA sponge.
Classic Case4
circRNA regulates its physiological functions by sequestering miRNA (ceRNA)
Paper: Zheng, Q., et al. (2016). "Circular RNA profiling reveals an abundant circHIPK3 that regulates cell growth by sponging multiple miRNAs." Nat Commun 7: 11215.
Circular RNA (circRNA) is a novel and prominent molecule in the field of ncRNA, and its functions are still poorly understood.
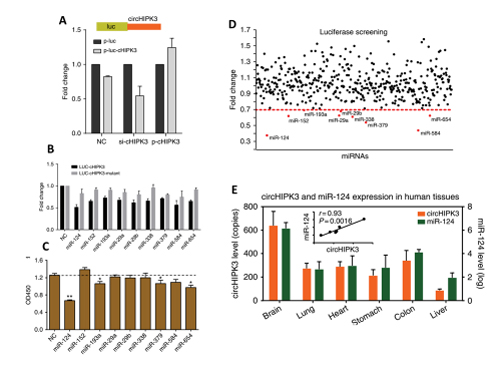
In this study, the authors identified a highly expressed circRNA molecule, circHIPK3, which originates from the second exon of the HIPK3 gene. RNA interference (RNAi) targeting circHIPK3 was found to affect cell growth. To further investigate its mechanism, the authors performed sequence analysis and discovered several miRNA binding sites within circHIPK3. In order to identify the miRNAs involved, the authors constructed a screening system based on luciferase activity. They cloned circHIPK3 into the 3'UTR region of Luciferase and co-transfected it with 424 miRNA mimics into the HEK-293T cell line for screening and validation. Ultimately, the authors identified 9 candidate miRNAs. Through functional experiments and co-expression analysis, mir-124 was confirmed as a positive candidate miRNA. In summary, the authors demonstrated that circHIPK3 positively regulated the expression of its target genes IL6R and DLX2 by adsorbing miR-124, thereby affecting cell growth.

Contact Hanbio and leave your requirements. We will reply as soon as possible.