- Lentivirus
- Adeno-Associated
- Adenovirus
- Pseudovirus
- Vector
- Synthesis
- Autophagy Research
- CRISPR/Cas9
- Noncoding RNA
- Luciferase Assay
- Reagents
WHAT ARE YOU LOOKING FOR?
Autophagy refers to the process by which non-essential or dysfunctional cellular components are degraded by lysosomes into essential nutrients, such as amino acids, lipids, and sugars. Autophagy serves not only as a specialized mechanism for cells to adapt to extreme environments but also as a crucial regulatory mechanism for normal cellular activities. Abnormal autophagy often plays a significant role in cell damage and aging. Currently, autophagy is known to be involved in various vital biological processes including tumor development, aging, inflammation, immune response, cardio-cerebral vascular diseases, oxidative stress, neurodegenerative diseases, metabolism, and development.
Autophagy refers to the process of non-specific degradation of cytoplasmic substrates such as proteins and organelles by hydrolytic enzymes within lysosomes. This process is finely regulated by complex signaling pathways involving PI3K-I/III and mTOR. The overall autophagic process can be divided into three stages: initiation stage of autophagy induction; formation stage of autophagosomes; and maturation stage leading to the formation of autolysosomes. Autophagy is implicated in important biological processes including tumor development, aging regulation, inflammation modulation, immune response, cardiovascular/cerebrovascular diseases, oxidative stress, neurodegenerative disorders, metabolism and development. In addition to studying autophagy itself as a fundamental process within cells, autophagy research is commonly investigated from mechanistic perspectives such as understanding its involvement in tumorigenesis progression, drug resistance, inflammatory responses, oxidative stress and the occurrence and treatment of autism and Alzheimer's disease.
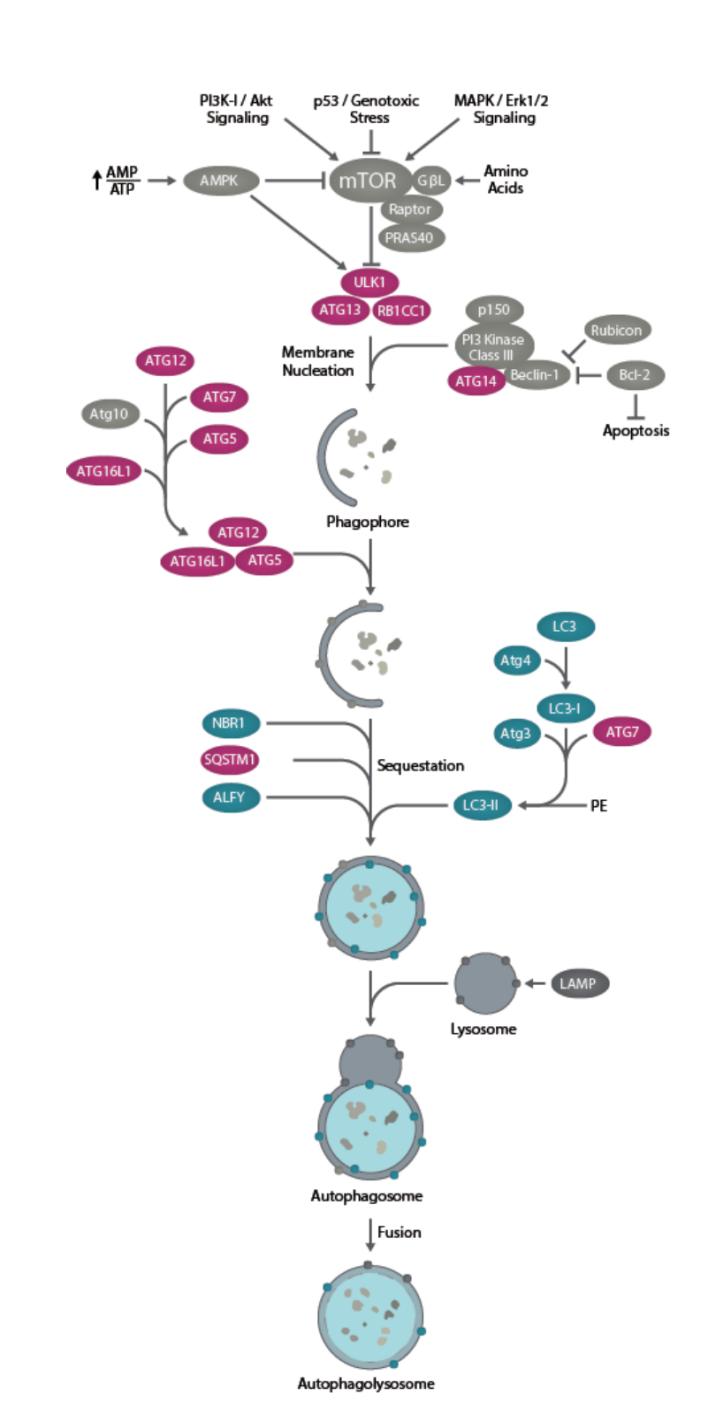
The Main Signaling Pathway and Related Genes of Autophagy
Product ID | Product Name | Production Scale |
HB-AP210 000 | dual-labeled fluorogenic LC3 adenovirus | 1mL (1010PFU) |
HB-LP210 0001 | dual-labeled fluorogenic LC3 lentivirus | 1mL (108TU) |
HB-AVP210 0001 | dual-labeled fluorogenic LC3 adeno-associated virus | 1mL (1012v.g) |
High authority: Hanbio dual-labeled fluorogenic LC3 adenovirus was published in the CELL journal.
Complete stock inventory: We have a comprehensive range of autophagy-related gene viruses and tool viruses in stock.
Numerous varieties: We offer a diverse selection of lentivirus, adenoviruses, adeno-associated viruses, and plasmid.
Monitoring changes in lc3 cleavage
Direct observation of autophagic vesicles using electron microscopy
Detection of autophagic flux using fluorescently-labeled lc3
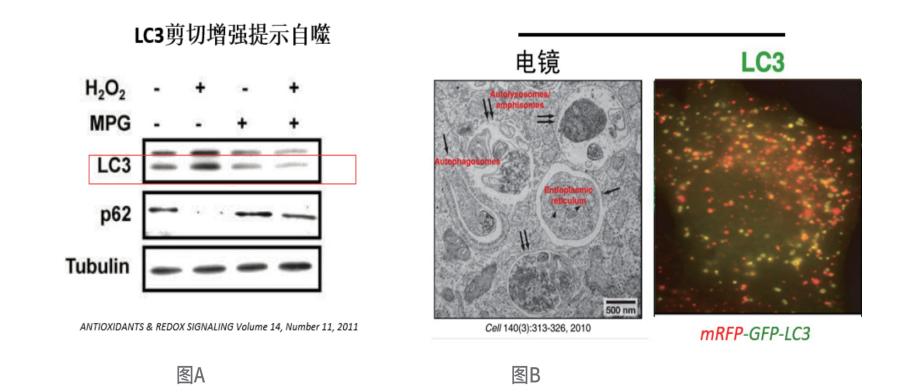
The three "golden standards" for assessing autophagy include post-translational processing and lipidation modification of LC3 (Figure A), direct observation of the number of autophagosomes and autolysosomes through electron microscopy, and monitoring changes in autophagic flux using fluorescently-labeled LC3 (Figure B).
Mainstream localization tracking tool one: mRFP-GFP-LC3 adenovirus (now mCherry-eGFP-LC3)
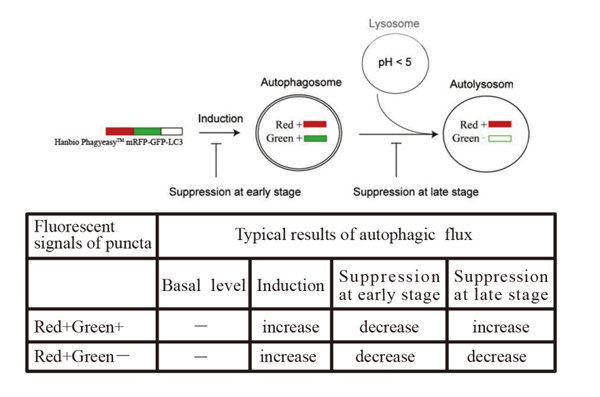
The principle of mRFP-GFP-LC3
mRFP enables tracking of LC3, while GFP reduction indicates fusion between lysosomes and autolysosomes forming autolysosomes. The principle of the assay is based on different pH stability of green and red fluorescent proteins. The fluorescent signal of EGFP could be quenched under the acidic condition inside the lysosome, and the mRFP fluorescent signal has no significant change under the acidic condition. In the autolysosomes stage, an acidic environment that quenches GFP fluorescence resulting only red fluorescence being detected.
In green and red-merged images, autophagosomes are shown as yellow puncta, while autolysosomes are shown as red puncta. The strength of autophagic flux can be clearly reflected by counting yellow and red punctas.
As depicted in the figure below, transfection of mRFP-GFP-LC3 virus into cells followed by amino acid deprivation treatment for 2 hours leads to a significant increase in autophagy and autophagic flux.
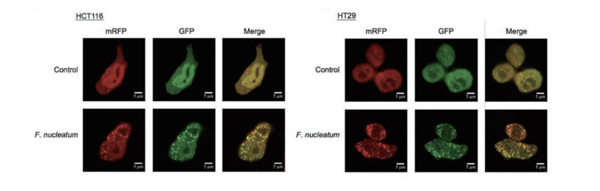
Red punctas without green indicate autolysosomes (mRFP), while yellow punctas represent autophagosomes (RFP+GFP). (Due to the sensitivity of GFP fluorescence protein to acidity, only red fluorescence can be detected when the autophagosome fuses with lysosome and GFP fluorescence is quenched.)
Fusobacterium nucleatum Promotes Chemoresistance to Colorectal Cancer by Modulating Autophagy.
This study was published by Hanbio customer in CELL in 2017 with an impact factor greater than 30.
Mainstream localization tracking tool two: p62
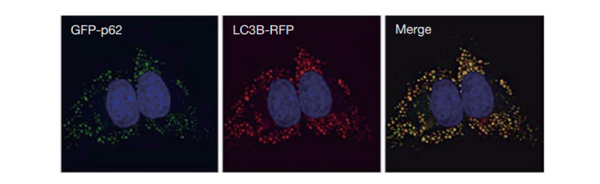
U2OS labeled with GFP-p62 and LC3-RFP simultaneously. Then treated with 60 μM chloroquine for 16 hours. Obvious colocalization between the two can be observed. (When autophagy is induced, GFP-p62 fusion protein is recruited to autophagosomes, appearing as green punctas. If chloroquine is added at this time to inhibit autophagic flux, then GFP-p62 will accumulate on both autophagosomes and autolysosomes)
Selective Autophagy Localization Tool: Keima probe
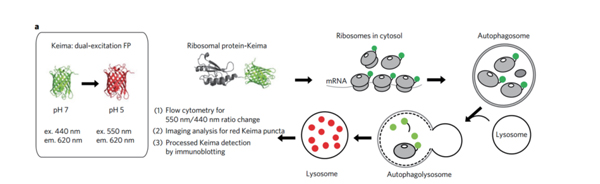
The basic principle of studying ribosomal selective autophagy using Keima probe
Fusobacterium nucleatum Promotes Chemoresistance to Colorectal Cancer by Modulating Autophagy
(journal: CELL, IF=41.582, Renji Hospital Affiliated to Shanghai Jiaotong University School of Medicine)
atg7-Based Autophagy Activation Reverses Doxorubicin-Induced Cardiotoxicity
(journal: Circulation Research, IF=17.367, Beijing University of Chinese Medicine)
PIWIL2 interacting with IKK to regulate autophagy and apoptosis in esophageal squamous cell carcinoma
(journal: Cell death and differentiation, IF=15.828, West China Hospital of Sichuan University)
USP11 regulates autophagy-dependent ferroptosis after spinal cord ischemia-reperfusion injury by deubiquitinating Beclin 1
(journal: Cell Death & Differentiation, IF=15.828, The First Affiliated Hospital with Nanjing Medical University)
Nanoprodrug ratiometrically integrating autophagy inhibitor and genotoxic agent for treatment of triple-negative breast cancer
(journal: Biomaterials, IF=12.479, The Second Affiliated Hospital Zhejiang University School of Medicine)

Contact Hanbio and leave your requirements. We will reply as soon as possible.